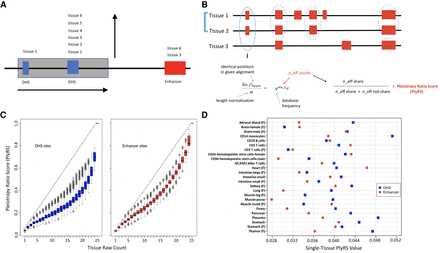
Pleiotropy ratio score (PlyRS). (A) Purifying selection against deletions can operate along two “axes” at a genomic locus. A deletion (here shaded gray) can remove putative genomic function along a horizontal axis by overlapping one or more regulatory sites (here overlapping two DHSs [hotspot; blue] but missing an enhancer [H3K4me1; red]). The regulatory sites overlapped can have annotated activity specific to a particular biosample (as the leftmost DHS) or cellularly pleiotropic activity (as the rightmost DHS), thus removing putative genomic function along a vertical axis. (B) Schematic of derivation of PlyRS. By using biosamples sharing an annotation at a given base pair, we compute effective number (n_eff) of biosamples by equating the observed sharing fraction to that under the expectation of independent annotations. PlyRS is given by n_eff values normalized across biosamples. (C) Comparison of PlyRS to simple biosample counts. PlyRS can range from zero, representing no regulatory activity at a particular base pair in any biosample, to one, representing regulatory activity in all biosamples analyzed. Because PlyRS accounts for the positive regulatory activity correlation between biosamples analyzed, PlyRS (y-axis) will always fall at, or below, the diagonal versus a simple count (x-axis). Each regulatory feature will display a different PlyRS distribution (e.g., the enhancer feature [H3K4me1; right] has a PlyRS distribution that lies closer to the diagonal than for the DHS feature [left]), based on the activity covariance of the biosamples that are analyzed of that regulatory feature. Boxplot midlines correspond to the median PlyRS value, with the box delimiting the second and third quartile range and the whiskers extending from box edges out up to 1.5 times the box range or the furthest PlyRS value if within that bound. (D) Comparison of PlyRS values for sites of activity specific to a particular biosample. When regulatory activity at a base pair is annotated as specific to a particular biosample, each biosample will have a PlyRS value corresponding to it that may be below, at, or above an otherwise simple count of 0.04 (i.e., one biosample divided by 25 total possible biosamples analyzed). This is owing to PlyRS up-weighting biosamples that have relatively rare activity genome-wide and down-weighting biosamples with relatively common activity. Because of the highly correlated nature of DHS biosamples (blue squares) to one another (or enhancer biosamples [red circles] to one another) among the biosamples that we analyzed, many biosamples fall below a count of 0.04 when regulatory activity at a base pair is specific to a particular biosample.











