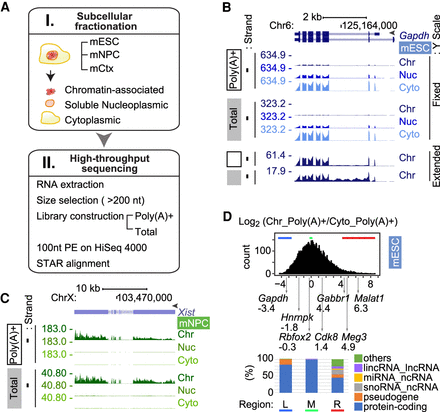
RNA partitioning between subcellular compartments. (A) Workflow used in this study. (B) Genome browser tracks of the Gapdh locus in mESCs. GENCODE annotated isoforms (M11) are diagrammed at the top. Poly(A)+ RNA (open box), total RNA (gray box), and peak RPM are noted on the left. RNA from chromatin (Chr), nucleoplasmic (Nuc), and cytoplasmic (Cyto) fractions are labeled at the right. The fixed Y-scale (RPM) shows the strong enrichment of Gapdh RNA in the cytoplasm. The bottom tracks show chromatin RNA with an extended Y-scale to observe the intron reads. (C) Genome browser tracks of the Xist/Tsix locus in female mNPCs show strong chromatin enrichment of Xist RNA. (D) Distribution of chromatin partition indices. The chromatin/cytoplasm ratio [Chr_Poly(A)+/Cyto_Poly(A)+] of the averaged read counts of each gene are plotted as a distribution along the log2 scale, with partition indices of representative genes indicated below. Biotypes of the 400 genes from bottom (left [L]; blue bar), peak (middle [M]; green bar), and top (right [R]; red bar) of the distribution are presented in the bar graph below.











