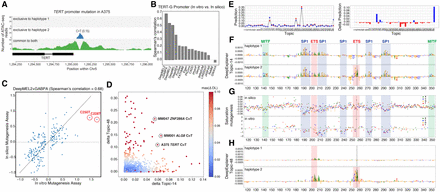
Analysis of TERT promoter mutations. (A) TERT promoter hotspot mutation in A375 is detected as an ASCAV as evidenced by ATAC-seq reads segregated into haplotypes (color-coded). In A375, haplotype 2 harbors the mutant allele T (according to WGS data) (see Supplemental Fig. S16), and ATAC-seq evidences exclusive accessibility for this allele. The corrected reference ATAC-seq allele ratio is indicated in parentheses. (B) Bar chart of model variant effect prediction performance on TERT promoter activity assessed by experimental saturation mutagenesis. (C) Scatter plot showing the effect of each variant in the in vitro (x-axis) and in silico (y-axis) mutagenesis of the TERT promoter. The two hotspot gain-of-function mutations are highlighted. (D) Scatter plot of delta Topic-14 score (promoter topic) versus delta Topic-48 score (GABPA topic) of all ASCAVs from 10 MM lines calculated by using the DeepMEL2 + GABPA model. ASCAVs are colored by their maximum delta prediction score. The TERT mutation of A375, as well as two newly predicted GABPA gains in MM047 and MM001 that are discussed in the text, are encircled. (E) The DeepMEL2 prediction score for each topic for both the haplotype 1 (red) and haplotype 2 (blue) of the A375 TERT locus is shown on the left, and the delta prediction scores between two haplotypes are shown on the right. The delta prediction scores for both Topic-14 (promoter topic) and Topic-48 (GABPA topic) are above the 0.05 detection threshold. (F,H) Haplotype-specific DeepExplainer plots of the A375 TERT promoter locus by using Topic-14 (F) and Topic-48 (H), annotated with the corresponding TFs. (G) Comparison of in silico (top; DeepMEL2 delta Topic-14 prediction scores) and in vitro (bottom; fold change in promoter activity) saturation mutagenesis assay. Each variant is color-coded.











