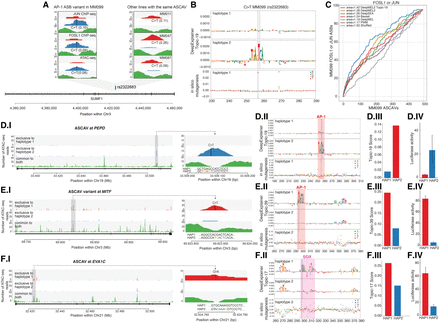
Model explanation and experimental validation of three cis-regulatory variants. (A) C > T intronic SNP (rs2322683) in SUMF1 is an ASCAV and AP-1 ASB (JUN and FOSL1 ChIP-seq data sets). (Left) Haplotypes 1 and 2 and unphased reads (color-coded) from this locus in MM099 JUN and FOSL1 ChIP-seq and ATAC-seq reads. (Right) Same locus in three additional MM lines (MM011, MM047, and MM087) in which rs2322683 is also inferred as an ASCAV. WGS genotypes (GT) and BaalChIP allele ratios are shown in parentheses. (B) DeepExplainer plot of the rs2322683 locus (position indicated with dashed lines), where the height of the nucleotides indicates their importance for the final prediction. Scoring using Topic-19 on both haplotypes shows C > T substitution generates an AP-1 binding site. In silico saturation mutagenesis on the reference sequence reveals the effect of each possible variant as a delta Topic-19 prediction score. (C) The curves represent the number of FOSL1 or JUN ASB variants found among the top-n MM099 ASCAVs ranked by the maximum delta prediction score of the different models. (D–F) Each row showcases the following: (I) an ASCAV and its allele-specific accessibility peak, (II) DeepExplainer and in silico mutagenesis results of the two haplotypes, (III) the DeepMEL2 score for both haplotypes, and (IV) the luciferase enhancer-reporter activity for both haplotypes. (D) C > T intronic variant in PEPD is identified as an ASCAV and predicted to generate an AP-1 binding site, with an increase in MES enhancer score. The in silico mutagenesis plot shows that only a single mutation to T at position 269 increases the MES enhancer prediction significantly, and this is exactly the location of the ASCAV. (E) C > T intronic variant in MITF is identified as an ASCAV and predicted to generate an AP-1 binding site. (F) G > A intronic variant in EVA1C is identified as an ASCAV and predicted to generate a SOX10 binding site.











