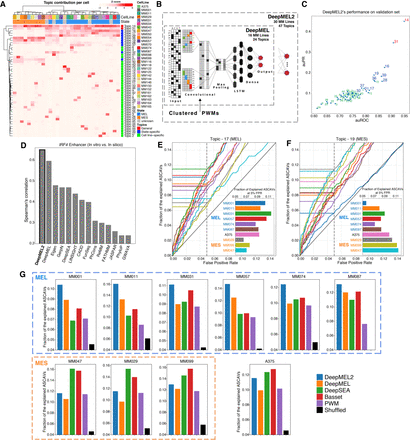
Cell state–aware DeepMEL2 can interpret ASCAVs. (A) Normalized cisTopic cell-topic heatmap of 30 melanoma cell lines showing general, state-specific, and cell line–specific sets of coaccessible regions. (B) Schematic overview of DeepMEL2 highlighting improvements compared with DeepMEL. (C) Scatter plot of auROC and auPR values shows the performance of DeepMEL2 on each topic. Promoter, state-specific, and cell line–specific topics are represented by red, blue, and green colors, respectively. (D) Performance of DeepMEL2 and other models at predicting variant effects on IRF4 enhancer activity. (E,F) Curves indicate fractions of ASCAVs explained by Topic-17 score (MEL; E) and Topic-19 score (MES; F) at different false-positive rates for each MM line. Bar chart insets show the exact fraction of the explained ASCAVs at 5% false-positive rate. (G) Bar charts showing the fraction of ASCAVs explained at 5% false-positive rate for each MM line using either DeepMEL2, DeepMEL, DeepSEA, Basset, and PWM. The black bar represents the fraction when ASCAVs and control variants are shuffled.











