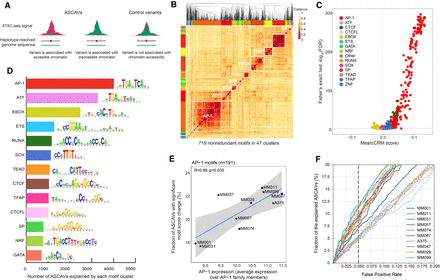
TF motif enrichment on ASCAVs. (A) Selection of ASCAVs and control variants used to assess the association between sequence content and allele-specific accessibility. (B) Heatmap showing the clustering of all 719 ASCAV-enriched motifs into 47 families (color-coded margins). The 13 major families are labeled with their cognate TF on the diagonal. (C) Scatter plot of motifs that are associated with chromatin accessibility. Each dot indicates a motif and is colored based on the motif cluster to which they belong. The x- and y-axes represent the delta cluster-buster motif score and the negative log-scaled FDR corrected P-value, respectively. (D) Bar plot showing the number of ASCAVs explained by each motif cluster. For each family, the consensus motif is shown. (E) Scatter plot of the average expression of AP-1 family members (JUN, JUNB, JUND, FOS, FOSB, FOSL1, FOSL2) and the fraction of ASCAVs that affects an AP-1 binding site. Correlation coefficient (Kendall's tau) and P-value are shown. (F) Fractions of ASCAVs explained at different false-positive rates are shown as curves for each MM line. Dashed lines represent the control for each MM line, where labels of ASCAVs and control variants are shuffled.











