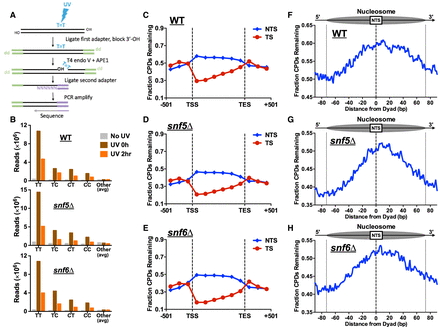
CPD-seq analysis indicates that SWI/SNF is dispensable for repair at most genomic and chromatin contexts. (A) Schematic of the CPD-seq method for mapping CPD lesions at single-nucleotide resolution. Adapted from Mao et al. (2016). (B) Enrichment of CPD-seq reads associated with putative CPD lesions at dipyrimidine sequences in UV-irradiated WT, snf5Δ, and snf6Δ cells but not the “No UV” control. (C–E) The fraction of CPDs remaining after 2-h repair was plotted for CPD-seq data for both the transcribed strand (TS) and nontranscribed strand (NTS) between the transcription start site (TSS) and transcription end site (TES) of approximately 5000 yeast genes. (F–H) Analysis of the fraction of CPDs remaining following 2-h repair within nucleosomes. Repair of the NTS oriented in the 5′ to 3′ orientation for the WT, snf5Δ, and snf6Δ cells was examined in aggregate for the first three nucleosomes (+1 to +3) downstream from the TSS of approximately 5200 genes. Dotted lines at positions −73 and +73 bp from the dyad center indicate the boundary of the nucleosome core particle.











