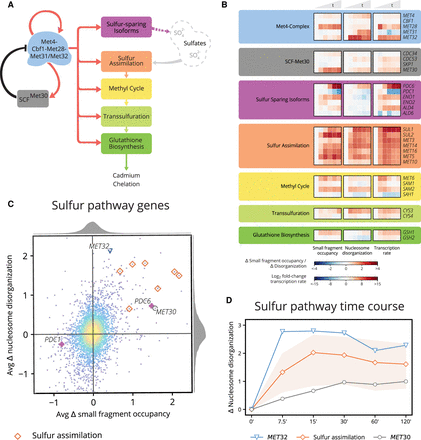
Chromatin and transcription dynamics detail Met4 and Met32 functional activation, induction of sulfur genes, and subsequent regulation. (A) The Met4 complex activates cascading sulfur pathways required for cadmium chelation and also activates its negative regulator SCFMet30. (B) Heat map of changes in chromatin occupancy and transcription rate for the sulfur pathway genes. Cofactors of the Met4 complex exhibit chromatin changes in small fragment occupancy (for MET28) and nucleosome disorganization (for MET32). Sulfur sparing isoforms occur as isoenzyme pairs; members of each pair exhibit inverse chromatin dynamics (most pronounced between PDC6 and PDC1). Nearly all of the sulfur assimilation pathway members show an increase in small fragment occupancy and nucleosome disorganization. (C) Scatterplot of average change in small fragment occupancy and average change in nucleosome disorganization. Chromatin dynamics in sulfur-related genes may manifest primarily in a single measure of the chromatin, as with MET32 (blue triangle), MET30 (gray circle), and PDC6/PDC1 (violet crosses), or in both small fragment occupancy and nucleosome disorganization, such as with the sulfur assimilation genes (orange diamonds). (D) Line plot of the change in nucleosome disorganization for the activator gene MET32, regulator gene MET30, and sulfur assimilation genes (orange line represents mean and light orange region represents full range of values across all seven genes). Nucleosome disorganization for Met4 complex cofactor MET32 peaks at 7.5 min whereas targets of the Met4 complex peak later: sulfur assimilation genes within 15–30 min and the negative regulator MET30 more gradually.











