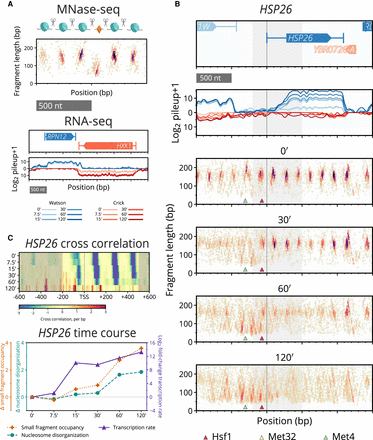
Paired-end MNase-seq and stranded RNA-seq capture high-resolution chromatin occupancy and transcriptome state throughout a perturbation time course. (A) Examples of MNase-seq and RNA-seq data. Top: Depiction of nucleosomes flanking a small (subnucleosomal) binding factor, and fragments that result upon digestion by MNase. Paired-end MNase-seq fragments appear in the typhoon plot based on their center position and length. Bottom: Strand-specific RNA-seq is plotted as the log of the pileup, the number of total RNA-seq reads at each genomic position, separately mapped to Watson (blue) and Crick (red) strands. RNA-seq levels over the time course are plotted using progressive coloring for each strand. (B) Beneath time course RNA-seq data, four typhoon plots show dynamics of MNase-seq data near HSP26 (gray shading highlights the [–200,+500] region around the TSS that we analyze for all genes). Nucleosomes in the promoter region are replaced by small fragments, and gene body nucleosomes disorganize. Small fragments appear around motifs for known regulators Hsf1 (red triangle), Met4 (green triangle), and Met32 (obscured by green triangle). (C) Plots of processed chromatin metrics around HSP26 over time. Top: Heat map of differential cross-correlation values through the time course, summarizing how gene body nucleosomes initially shift downstream and eventually disappear, and how promoter nucleosomes are rapidly displaced as small fragments accumulate. Higher values (more red) indicate higher cross-correlation with subnucleosomal fragments; lower values (more blue) indicate a stronger signal for nucleosomal fragments. Bottom: Line plot summarizing the change in occupancy of promoter small fragments (orange), disorganization of gene body nucleosomes (turquoise), and transcription rate (purple) of HSP26 over the time course.











