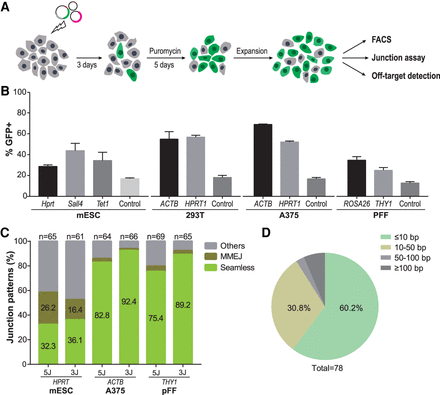
Systematic assessment of the HIT-trapping method in mammalian cells. (A) Flowchart of the experimental design. (B) HIT-trapping efficiencies were measured by the percentage of GFP-positive cells among puromycin-resistant cells. As controls, plasmid mixtures without gene-specific sgRNA expressing vectors were transfected into the cells. These results were derived from at least two independent experiments, shown as mean ± SD. (C) Frequencies and ratios of seamless, microhomology-mediated end joining (MMEJ), and other integration junctions at the target loci. (n) Number of the amplicons analyzed; (5/3J) 5′/3′ junction. (D) Summary statistics of the deletion sizes from junction sequencing. Sequencing data are presented in Supplemental Figure S6.











