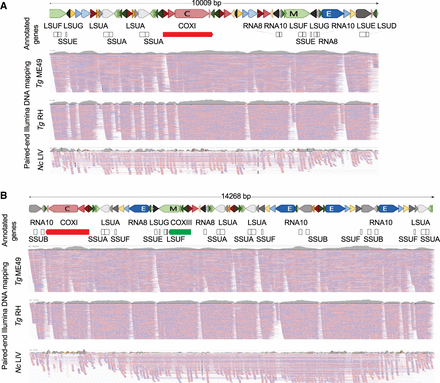
ONT and Illumina comparisons reveal sequence block order variability and decay with evolutionary time. (A,B) Two T. gondii ME49 ONT sequence reads were annotated with the gene sequences they contain. Reads and SBs are drawn to scale. The blocks are colored as shown in Table 1. “Annotated genes” track represents the annotation of the cytochrome genes and rRNA gene fragments on the ONT reads. The three “PE Illumina DNA mapping” tracks show PE read mapping of T. gondii ME49, TgRH88, and N. caninum (Nc) LIV mtDNA-specific reads. TgME49 and TgRH reads mapping required 100% nucleotide identity whereas 1% mismatch was allowed for mapping NcLIV reads. Reads were independently mapped to each of the ONT mtDNA reads and visualized using IGV. Red and blue lines below each read indicate the mapped Illumina PE reads. Coverage plots of the mapped reads are shown above each mapping panel in gray to indicate the depth of all reads and not just those shown.











