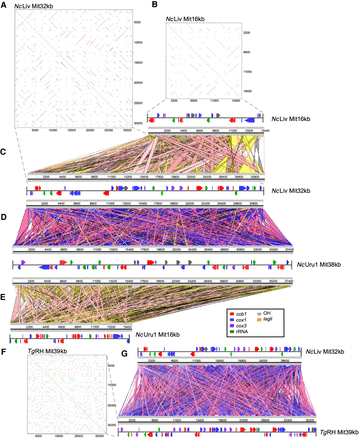
Comparative analysis of mitochondrial genome structures and annotations of Neospora and Toxoplasma reveals gene fragmentation and reshuffling between species and strains. (A) The repetitive nature of the gene structure in a 32-kb mitochondrial DNA contig of NcLiv is graphically represented in a YASS plot. (B) The repetitive nature of the gene structure in a 16-kb mitochondrial DNA contig of NcLiv is graphically represented in a YASS plot. (C) Comparative alignment between two NcLiv mitochondrial contigs of 16 and 32 kb, respectively. (D) Comparative alignment between a NcLiv mitochondrial contig of 32 kb and a NcUru1 mitochondrial contig of 38 kb. (E) Comparative alignment between two NcUru1 mitochondrial contigs of 16 and 38 kb, respectively. (F) The repetitive nature of the gene structure in a 16-kb mitochondrial DNA contig of NcUru1 is graphically represented in a YASS plot. (G) Comparative alignment between a NcLiv mitochondrial contig of 32 kb and a T. gondii mitochondrial contigs of 39 kb.











