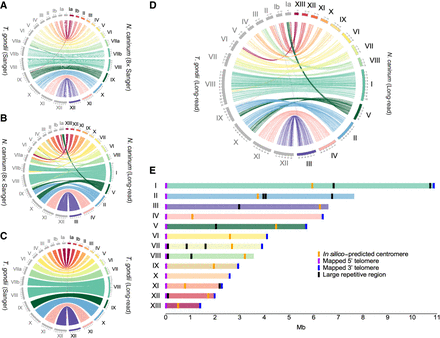
Comparative analysis of genome assemblies of Neospora caninum and Toxoplasma gondii using third-generation sequencing data reveals misassembly and karyotype differences. (A) Comparative analysis of the T. gondii type II (TgME49) genome assembly and the N. caninum Liverpool (NcLiv) strain genome assembly, obtained based on Sanger technology sequencing data. (B) Comparative alignment of the NcLiv genome assemblies using Sanger and third-generation (long-read) technology. (C) Comparative alignment of the T. gondii type II (TgME49) genome assemblies based on Sanger technology sequencing data or third-generation (long-read) technology of T. gondii type I (TgRH). (D) Comparative alignment of the T. gondii type I (TgRH) and the NcLiv genome assemblies based on third-generation (long-read) sequencing technology. (E) Chromosomal layout of N. caninum. Karyotype, chromosome length, telomeres, putative centromeres, and large repeats are shown.











