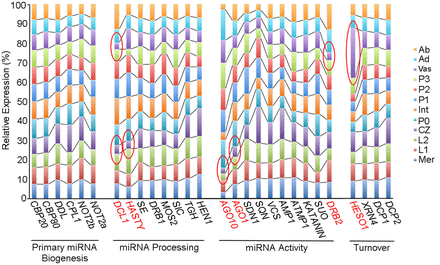
Relative expression of key miRNA pathway genes varies across shoot apex domains. The bar graph illustrates the relative expression of miRNA pathway functions across the 12 apex domains. Although transcript levels for most genes change minimally across the apex, DCL1, HASTY, AGO1, AGO10, HESO1, and DRB2 show strong expression variation, particularly in the vasculature and CZ (highlighted red). For functions represented by multiple paralogs in the maize genome, the bar graph is based on cumulative expression of all paralogs (Supplemental Data Set S2).











