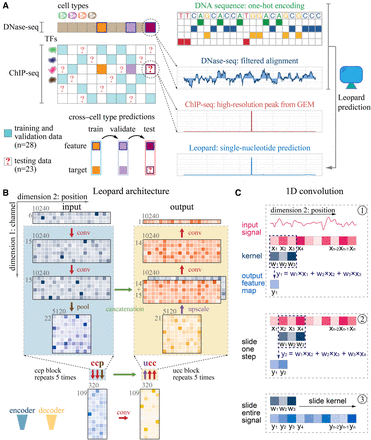
Schematic illustration of Leopard workflow. (A) This study aims to decode the high-resolution transcription factor (TF) binding landscapes (ChIP-seq peaks extracted by GEM peak finder) based on chromatin accessibility (DNase-seq signals) in a cross–cell type fashion. A total of 28 ChIP-seq experimental results from the ENCODE Project were used to train and validate models, whereas the other 23 results were used to test the performance of our method. The DNase-seq signals of filtered alignments and one-hot-encoded DNA sequences were used as inputs for a deep convolutional neural network model. (B) Leopard accepts two-dimensional matrices as inputs, where the first dimension represents six channels (DNase-seq, ΔDNase-seq, and one-hot-encoded DNA sequence) and the second dimension represents 10,240 genomic positions. The 10,240 genomic positions correspond to randomly sampled consecutive segments in the human genome. Leopard has two components: the encoder (blue) and the decoder (yellow). The encoder contains five convolution-convolution-pooling (ccp) blocks, and the decoder has five upscaling-convolution-convolution (ucc) blocks. This architecture allows for generating outputs for multiple positions simultaneously, substantially boosting the prediction speed. In addition, the concatenation operations (horizontal green arrows) connect the encoder with the decoder, preventing information decay in deep neural networks. (C) The one-dimensional (1D) convolution operator calculates the inner product between the kernel (w1, w2, w3) and the input signal (x1, x2, x3), resulting in one feature map value (y1) in step 1. Then the kernel slides along the entire input signal (steps 2 and 3) and generates the output feature map vector, which has the same size in dimension 2 as the input.











