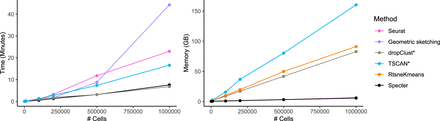
Runtime and peak memory usage as a function of sample size. Seurat was run with a call to the more efficient scanpy implementation of the Louvain clustering algorithm. Running times exclude preprocessing for all methods except TSCAN and dropClust, whose implementation did not allow us to isolate the core algorithm. Memory usage of Specter, Seurat, and geometric sketching are nearly identical and cannot be distinguished in this plot. For ease of visualization, we show runtime results of method RtsneKmeans in Supplemental Figure S21.











