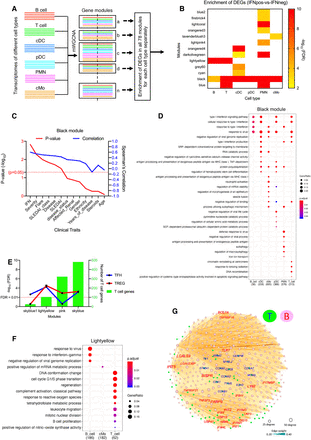
Multi–cell type WGCNA (mWGCNA) analysis reveals IFN-driven cross-talk between T cells and B cells. (A) A figure depicting the mWGCNA using combined transcriptomes from six different cell types. The mWGCNA has generated 78 different modules, and example modules show different proportion of genes from six different cell types. (B) We found 13 modules (out of total 78) that have significant enrichment of DEGs (IFNpos-vs.-IFNneg) in at least one cell type. The x-axis shows modules with their enrichment in each cell type and y1-axis shows −log10 of the adjusted P-value (based on a hypergeometric test). This includes four modules (black, blue, light yellow, and dark olive green) where the DEGs are significantly enriched in two or more cell types. (C) The line plot shows significance (−log10 of P-value) on the y1-axis in red and Spearman's correlation on the y2-axis in blue for the correlation of different traits with the black module eigengene. The clinical traits are sorted in decreasing order of statistical significance of the correlation for the black module. (D) Functional annotations (generated by clusterProfiler) of genes from each cell type in the black module. The color shows the significance (adjusted P-value), and the size of the dot corresponds to the proportion of genes with the corresponding annotation. (E) Enrichment of TFH and TREG gene sets in all 78 modules highlighting four modules with significant overlap for at least one of these gene sets. The x-axis is different modules, the y1-axis is −log10 of the adjusted P-value (based on a hypergeometric test), and the y2-axis shows the number of genes from T cells present in the corresponding module. (F) The functional annotations of significantly enriched genes from different cell types in the light yellow module. (G) Examples of T cell and B cell genes from the light yellow module as visualized by Gephi. The nodes are colored according to the cell of origin (T cells in green node with blue text and B cells in pink node with red text) and sized according to the number of edges (connections), and the edge thickness is proportional to the strength of coexpression.











