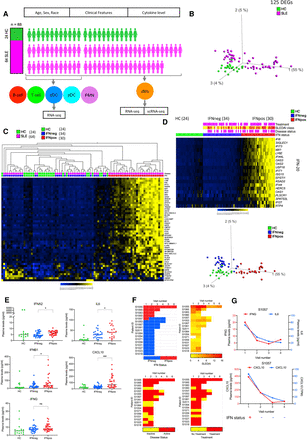
Transcriptional profiling of classical monocytes reveals two molecular subtypes of SLE. (A) Overview of the SLE cohort. Healthy control (HC) samples are highlighted in green color and SLE samples are highlighted in magenta color. Six different immune cell types, including classical monocytes (cMo), Polymorphonuclear Neutrophils (PMNs), conventional dendritic cells (cDC), plasmacytoid dendritic cells (pDC), T cells, and B cells are displayed in different colors. (B) The PCA plot based on 125 DEGs (P.adj < 0.05 from a Benjamini–Hochberg test in DESeq2) between SLE and HC in classical monocytes. Green and magenta colors represent HC and SLE patients, respectively. (C) Heat map of top 50 most variable DEGs (one per row) in a SLE versus HC comparison (P.adj < 0.05) clustered with hierarchical clustering and presented as row-wise z-scores (−2.0 [blue] to 2.0 [yellow]) of transcripts per million (TPM) in SLE (magenta) or HC (green); each column represents an individual patient. IFNpos (red) and IFNneg (blue) are shown in different colors. (D) SLE patients (first study visits) within each group (HC, IFNneg, IFNpos) are sorted with respect to the first principal component of IFN-20 gene expression. The SLEDAI CLASS panel shows the SLEDAI score and is divided into three different categories based on score: DA1 (0–2), DA2 (3–7), and DA3 (>7) (yellow, orange, and red, respectively). Patients with active disease status and those taking treatment are highlighted in purple color. This color scheme is used throughout all figures. (E) Scatter dot-plots show the expression level (pg/ml) in plasma of IFNG, IL6, and CXCL10. Only first visit samples (n = 76) were used in these scatter dot-plots. The difference in measurements from IFNpos and IFNneg patients has been calculated using unpaired t-test, two-tailed. (ns) Not significant, (*) <0.05, (***) <0.001. (F) Longitudinal IFN response status of 17 SLE patients (five patients were IFNpos and 12 patients were IFNneg at the first study visit) (top left). Only one patient (S1057) changed their IFN response status between study visits, changing from IFNpos to IFNneg between the first and second study visit and remained IFNneg in all follow-up visits. Similar heat maps of IFN status and different clinical parameters such as SLEDAI, disease status, and treatment for patients with longitudinal information. (G) Two plots with connecting lines (upper and lower) show expression changes of multiple cytokines and chemokines in the longitudinal data for patient S1057, where different analytes are plotted on different y-axes (IFNG on y1-axis in red color and IL6 on y2-axis in blue color) in different colors. The RNA-level expression of CXCL10 is also shown (y2-axis) with protein-level expression of CXCL10 (y1-axis). The lower panel of each plot shows their IFN response status over multiple longitudinal visits.











