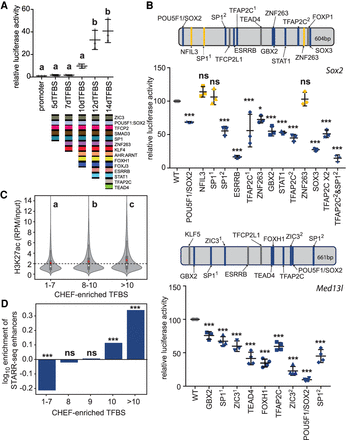
Ten or more different transcription factor binding sequences are required for enhancer activity. (A) Sequential removal of TFBS to form 14dTFBS_a revealed the importance of multiple TFBS sequences for enhancer activity and a threshold requirement of 10 TFBSs. Error bars represent the standard deviation; groups determined by one-way ANOVA to be significantly different (P < 0.05) are labeled with different letters. (B) Enhancers that regulate Sox2 or Med13l contain multiple conserved TFBSs (top) which are required for activity as demonstrated by TFBS mutagenesis (bottom). TFBSs indicated in blue were required for enhancer activity; TFBSs indicated in gray were not modified; yellow indicates TFBSs found not to be required for activity. Significant differences compared to the wild-type (WT) sequence are indicated by (*) P < 0.05, (***) P < 0.001, (ns) = not significant. Error bars represent the standard deviation. (C) Transcription factor–bound regions in mouse ESCs with >10 TFBSs have significantly higher enrichment of H3K27ac compared to transcription factor–bound regions with 8–10 and 1–7 TFBSs in the 700-bp sequence window. Groups determined by one-way ANOVA to be significantly different (P < 0.0001) are labeled with different letters. In the violin plots, the white dot indicates the median, the red box indicates the mean, and the dashed line indicates the average for the 1–7 TFBSs group. (D) Regions containing 10 or >10 CHEF-enriched TFBS sequences have significant enrichment of STARR-seq enhancers as indicated by (***) (P < 0.001, hypergeometric test). Regions containing 1–7 CHEF-enriched TFBS sequences are depleted in STARR-seq enhancers; (ns) indicates no significant enrichment of STARR-seq enhancers.











