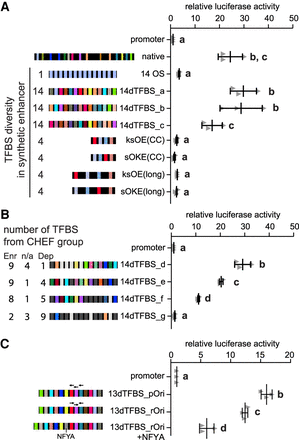
Synthetic sequences reveal that transcription factor binding site diversity is required and sufficient for robust enhancer activity. In panels A–C, error bars represent the standard deviation; groups determined by one-way ANOVA to be significantly different (P < 0.05) are labeled with different letters; to indicate P > 0.05, groups are labeled with the same letter. n ≥ 3 biological replicates. (A) Synthetic enhancers were evaluated in reporter assays and compared to the activity of the Sox2 enhancer (native). A sequence containing 14 POU5F1:SOX2 TFBS (14OS), 14 different TFBSs (14dTFBS_a, _b, _c) from the CHEF-enriched TFBS were evaluated. Optimized 4 TFBS sequences (ksOE, sOKE), with either CC or long spacers between motifs were also evaluated. (B) Enhancer activity is reduced when sequences contain fewer CHEF-enriched (Enr) and more CHEF-depleted (Dep) TFBSs. The number of TFBSs that are neither enriched or depleted is indicated by n/a. (C) The effect of motif orientation and repressor binding on enhancer activity. 13dTFBS_pOri contains the preferred orientation, 13dTFBS_rOri contains reversed TFBS. Addition of the repressor NFYA to 13dTFBS_rOri affects but does not abolish enhancer activity.











