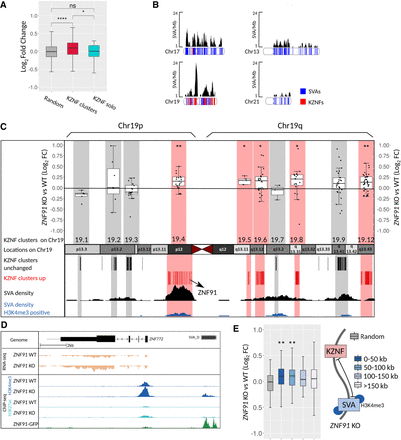
Up-regulation of clustered KZNF genes in proximity of activated SVA elements. (A) Box plots showing log2 fold change of a random gene set (257), KZNF located in clusters (265), and solitary KZNF genes (38). Clusters and solitary KZNF were defined by Thomas and Schneider (2011). Only genes with a baseMean > 10 were included. (B) SVA coverage (SVA/Mb) is shown for Chromosomes 17 and 19 with higher SVA density than expected and Chromosomes 13 and 21 with lower SVA density than expected (based on Wang et al. 2005). Chromosome density plots show distribution of SVA elements (blue) and KZNF genes (red). (C) Schematic representation of Chromosome 19 showing the location of KZNF clusters. Box plots of log2 fold changes on top show if clusters are up-regulated (red) or unchanged (gray). Only genes with baseMean > 10 were included. Number of expressed genes per cluster: 19.1 = 9 genes, 19.12 = 42 genes, 19.2 = 7 genes, 19.3 = 20 genes, 19.4 = 23 genes, 19.5 = 5 genes, 19.6 = 24 genes, 19.7 = 3 genes, 19.8 = 19 genes, 19.9 = 26 genes. Coverage tracks show SVA/Mb for all SVA elements (black) and H3K4me3-positive SVA elements (blue). (D) Coverage of RNA-seq and ChIP-seq (H3K4me3 and H3K27ac) on ZNF91 WT and KO hESCs and ZNF91 in HEK293 cells showing ZNF772 and upstream SVA-D. RNA-seq coverage tracks were scaled using DESeq2 scaling factors. ChIP-seq coverage tracks were scaled based on control regions. (E) Box plots showing log2 fold change of KZNF grouped per distance to nearest SVA that gained H3K4me3 upon ZNF91 deletion (random = 88 genes, 0–50 kb = 112 genes, 50–100 kb = 60 genes, 100–150 = 46 genes, >150 kb = 85 genes). For all analyses only expressed (baseMean > 0) genes were included. For all statistical testing an unpaired Wilcoxon rank-sum test was used, followed by an FDR correction: (ns) P > 0.05; (*) P < 0.05; (**) P < 0.01; (****) P < 0.0001.











