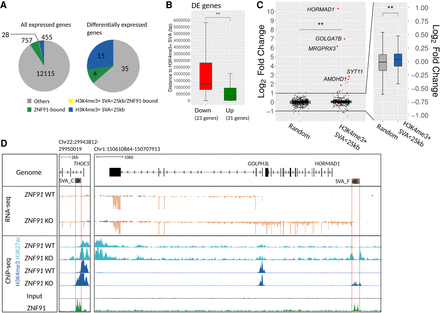
Differentially expressed genes in ZNF91 KO cells are bound by ZNF91 and driven by activated SVA elements. (A) Pie charts of all expressed (baseMean > 10) and differentially expressed (DE) genes (DESeq2 analysis, Padj < 0.05) indicating how many genes are bound by ZNF91 at their promoter (green), located within 25 kb of an H3K4me3-positive SVA (blue), both (yellow), and neither (gray). (B) Box plots of distance to nearest H3K4me3-positive SVA from TSS of DE genes (23 genes down-regulated and 31 up-regulated). (C) Box plots showing log2 fold change of genes that are located within 25 kb of an H3K4me3-positive SVA (420 genes) compared to a set of randomly selected genes (556 genes). Red data points indicate genes that were differentially expressed (FDR < 0.05). Only expressed genes (baseMean > 10) were included in this graph. (D) Coverage tracks of RNA-seq (blue is transcription from the positive strand, and orange is from the negative strand) and ChIP-seq data: (left) activation of an intronic SVA-C results in extension of H3K4me3 and H3K27ac signals at the promoter, leading to increased expression of THOC5; (right) activation of an SVA-F upstream of HORMAD1 leads to generation of an SVA-HORMAD1-GOLPH3L fusion transcript. Red boxes indicate activated SVA elements. RNA-seq coverage tracks were scaled using DESeq2 scaling factors. ChIP-seq coverage tracks were scaled based on control regions. For all statistical testing, an unpaired Wilcoxon rank-sum test was used, if necessary, followed by an FDR correction: (**) P < 0.01.











