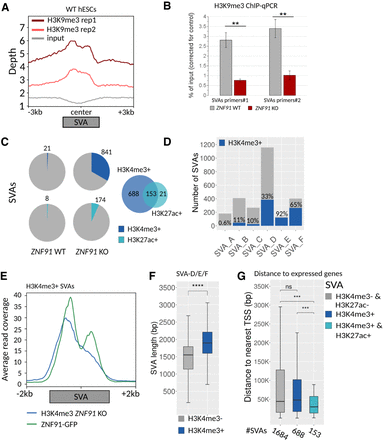
Evolutionarily young SVA elements become epigenetically active in the absence of ZNF91. (A) Profile plots showing coverage of H3K9me3 centered on SVA elements in WT hESCs, and 3 kb upstream of and downstream from SVAs is displayed. (B) H3K9me3 ChIP-qPCR on ZNF91 WT and KO hESCs using two SVA primer pairs. Percentage of input was normalized against a control (LTR12). A one-sided, unpaired t-test was used for statistical analysis. (C) Pie charts showing number of H3K4me3 (dark blue) and H3K27ac (light blue) positive SVA elements in ZNF91 WT and KO hESCs; Venn diagram showing overlap of H3K4me3 and H3K27ac peaks. (D) Absolute number (gray) and percentage of H3K4me3-positive (blue) SVA elements in each subclass. (E) Profile plot showing ZNF91 ChIP signal in HEK293 cells and H3K4me3 signal in ZNF91 KO hESCs. Average read coverage on H3K4me3-positive SVAs was displayed 2 kb upstream of and downstream from the element. (F) Box plot showing length of H3K4me3-positive and -negative SVA subtypes D, E, and F. (G) Box plots showing distance from H3K4me3/H3K27ac-negative, H3K4me3-positive, and H3K4me3/H3K27ac-positive SVA elements to nearest TSS of a gene expressed in hESCs (baseMean > 10) in base pairs. Outliers are not displayed. For statistical testing in F and G, an unpaired Wilcoxon rank-sum test was used, for G followed by an FDR correction: (****) P < 0.0001; (***) P < 0.001; (**) P < 0.01; (ns) not significant. For H3K4me3 and H3K27ac ChIP analyses, only SVA elements bound by ZNF91 were included.











