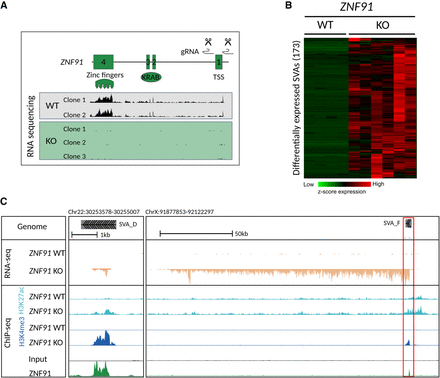
SVA elements become transcriptionally active in the absence of ZNF91. (A) CRISPR-Cas9 design to delete 172 bp around the transcription start site (TSS) of ZNF91 and RNA-sequencing coverage tracks showing absence of ZNF91 expression in ZNF91 KO clonal hESC lines. (B) Heatmap of hierarchical clustering (Euclidean distance) using Z-score of scaled counts of SVA elements that are up-regulated in ZNF91 KO hESCs (defined as log2 fold change ≥ 3). (C) Coverage of RNA-seq and H3K4me3 and H3K27ac ChIP-seq data for ZNF91 WT and KO hESCs and ZNF91-GFP ChIP on HEK293 cells: (left) transcripts running from 5′ to 3′ end of SVA; (right) red box indicates SVA element as driver of lncRNA. RNA-seq coverage tracks were scaled using DESeq2 scaling factors. ChIP-seq coverage tracks were scaled based on control regions.











