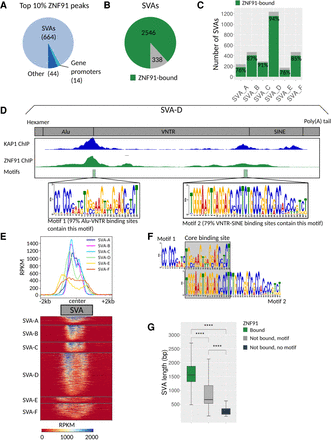
ZNF91 binds at the Alu-VNTR and VNTR-SINE border of all SVA subclasses. (A) Pie chart showing the distribution of the top 10% ZNF91 peaks based on MACS score. (B) Pie chart showing the number of SVA elements that are bound (green, 2545) and not bound (gray, 338) by ZNF91. (C) Frequency plot showing fraction of SVA subclasses bound by ZNF91 (green). (D) Schematic of SVA-D with different domains indicated. A pile-up of KAP1 summits on the SVA-D consensus is displayed in blue (Jacobs et al. 2014), and a representative example of ZNF91 coverage on an SVA-D is in green. Two ZNF91 binding motifs were identified localizing to the Alu-VNTR border and VNTR-SINE border. The fraction of SVA elements that contain motif 1 in the peaks localizing to the Alu-VNTR border and motif 2 in the peaks localizing to the VNTR-SINE border are expressed in percentages. (E) Heatmap and profile plots of ZNF91 ChIP showing coverage in RPKM of the mean of replicates 1 and 2 with SVA center as reference point. Average signal plotted per SVA subclass. (F) Comparison of motifs 1 and 2 showing overlap at the center. (G) Length of SVA elements bound (2546, green) and not bound (gray) by ZNF91. Light gray indicates that at least one of the two SVA binding motifs (199) was present, and dark gray indicates that no motif was detected (139). For statistical testing, an unpaired Wilcoxon rank-sum test was used followed by an FDR correction: (****) P < 0.0001.











