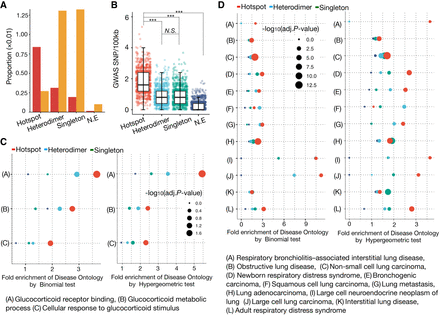
Disease-associated genetic variants and GO enrichment in distinct AP-1 binding classes. (A) Bar plot showing the proportion of each AP-1 binding class that contains at least single GWAS SNP. (B) Box plot showing the distribution of trait-associated SNP density (SNP/100 kb sequence) for distinct AP-1 binding classes. Each dot represents the SNP density based on the same number of loci repeatedly subsampled from each class. P-values were calculated using Wilcoxon rank-sum test. (***) P < 0.001, N.S. = P > 0.05. (C) GO enrichments using the genomic region-based binomial test (left) and gene-based hypergeometric test (right) for distinct AP-1 binding classes. The color and size of dots represent the different AP-1 binding classes and the adjusted P-values for a given annotation, respectively. (D) Disease Ontology (DO) enrichments using the genomic region-based binomial (left) and gene-based hypergeometric test (right) for distinct AP-1 binding classes. The color and size of dots represent the different AP-1 binding class and the adjusted P-value for a given annotation, respectively.











