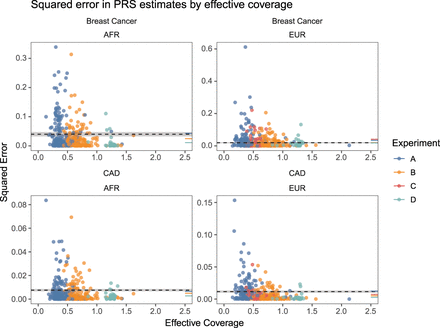
For each imputed sample, we computed a PRS for breast cancer and CAD and calculated the squared error of the estimate compared to the PRS of the “truth” genotypes in the 1KGP3. Each dot represents this squared error for each sequenced sample for a given trait and is colored by the experiment to which it belongs. To provide a point of comparison to PRS estimated from imputed array data, we computed the mean squared error for each cell line across array replicates and averaged that across all cell lines. This quantity is represented by the dashed line for each trait and superpopulation along with the standard error of the mean, represented by the shaded regions about each dashed line. The mean squared error for each experiment was calculated in the same way and is rendered as a colored line segment on the rightmost margin of each panel. These results indicate that sequencing at effective coverages of 0.5 or higher generally affords lower measurement error in PRS estimates.











