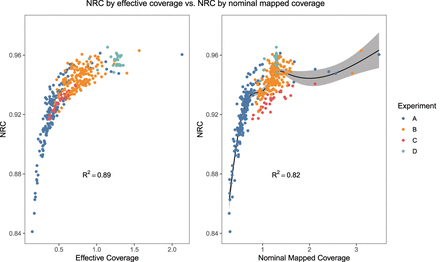
Nonreference concordance (NRC) (Methods) for imputed SNPs for all EUR samples in experiments A–D plotted against effective coverage λeff (left) or nominal mapped coverage (right). We modeled the NRC as the response variable for a k = 5 knot cubic restricted spline with the respective coverage metric as the explanatory variable; the fitted values are shown as a solid line with the surrounding shaded regions representing 95% confidence intervals. The knot locations were set at the 5th, 27.5th, 50th, 72.5th, and 95th percentiles of the respective coverage metrics following Harrell's rule of thumb (Harrell 2017).











