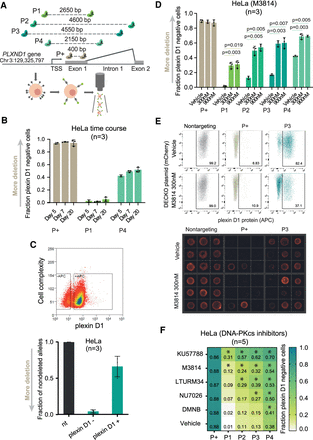
CiDER reporter system identifies DNA-PKcs inhibition as a means to increase CRISPR-del efficiency. (A) The CiDER endogenous reporter relies on a series of sgRNA pairs targeting exon 1 of PLXND1 gene, whose protein product is detected by flow cytometry. (B) CiDER measurements of CRISPR-del efficiency over a range of times between sgRNA delivery and cell harvesting (mean, standard deviation). (C) Independent evaluation of CRISPR-del efficiency in sorted plexin D1–negative and D1–positive cells. (Top) Representative fluorescent activated cell sorting (FACS) plot. Plexin D1–negative (APC-) and Plexin D1–positive cells (APC+) are gated. (Bottom) The fraction of nondeleted alleles in each population quantified by qPCR (mean, standard deviation). The black bar corresponds to a nontargeting control used for normalization. Green bars correspond to sorted HeLa cells with pgRNAs targeting PLXND1 (P3). (D) CRISPR-del efficiency measured by CiDER in HeLa upon DNA-PKcs inhibition (mean, standard deviation, one-tailed paired t-test). (E, top) Representative raw flow cytometry plots for HeLa upon DNA-PKcs inhibition. Plexin D1–positive cells are gated, and numbers correspond to percentage of cells. (Bottom) Representative images of individual sorted, stained HeLa cells. (F) CiDER measurements of CRISPR-del efficiency in HeLa upon DNA-PKcs inhibition with indicated small molecules (values: mean plexin D1–positive cells; *P < 0.05, one-tailed paired t-test).











