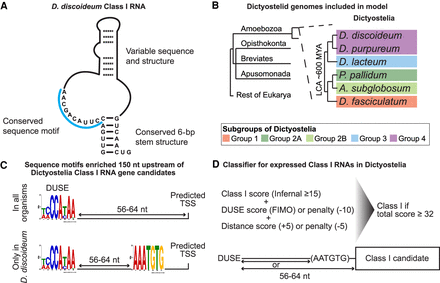
Search strategy and classification of Class I RNAs. (A) Schematic representation of previously described D. discoideum Class I RNAs (Avesson et al. 2011). (B) Schematic phylogeny showing the location of Amoebozoa, a sister group to Obazoa (Opisthokonta, Breviates, and Apusomonada) in the eukaryotic tree of life based on Burki et al. (2020). Dictyostelia is represented by species belonging to each major group (Schilde et al. 2019). The genomes of these dictyostelids were searched for Class I RNA genes, and newly identified genes were used to refine the covariance model. (C) Enriched sequence motifs identified upstream of Class I RNA gene candidates in the different dictyostelids represented in B (Infernal score ≥ 25, n = 126). The putative promoter motif (DUSE) is found ∼60 nt from the predicted start of transcription (TSS) in all organisms (upper). DUSE in combination with TGTG box, only identified in D. discoideum (lower). (D) Summary of scoring system used for the classifier of Dictyostelia Class I RNA based on Infernal score ≥ 15, presence of DUSE, and distance between DUSE and predicted TSS or TGTG box.











