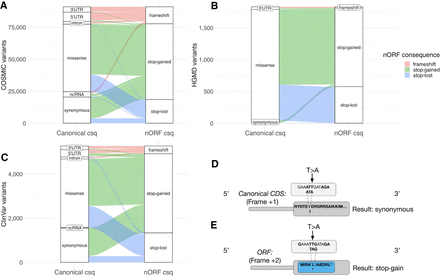
Reinterpreting COSMIC, HGMD, and ClinVar mutations in the context of nORFs. The canonical consequence and nORF consequence of (A) 109,000 somatic cancer mutations from COSMIC, (B) 1852 disease mutations from HGMD, and (C) 5269 disease mutations from ClinVar. Bins with 10 or fewer variants are not shown. These mutations would likely be interpreted as benign or missense in canonical genes but may have more severe consequences in nORFs. (D) A theoretical example of a disease variant that results in a synonymous mutation in canonical CDS but a stop-gain mutation in a nORF from an alternative reading frame (E).











