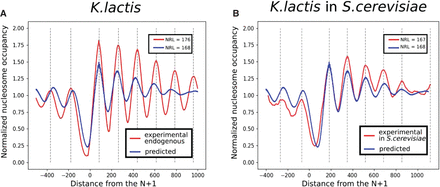
Predictions on the Kluyveromyces lactis genome compared with experiments. The prediction of the model, trained on Saccharomyces cerevisiae and applied on Chromosome F of K. lactis, is compared with the experimental data obtained by Hughes et al. (2012). The signal around every TSS is aligned with respect to the first nucleosome downstream from the TSS and averaged. (A) Endogenous context (Chr F of K. lactis in K. lactis). (B) Transfected context (Chr F of K. lactis in S. cerevisiae).











