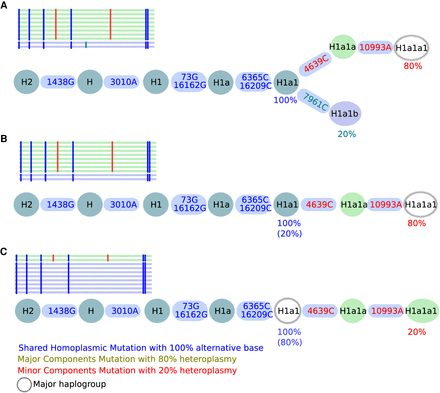
All possible contamination scenarios. Here, a contamination level of 20% is shown in all three scenarios (A–C). Shared polymorphisms of two haplotypes are included in a single branch, whereas the split into two branches displays the different lineage haplotypes. (A) Shared mutations defining H1a1 (last common ancestor [LCA]) are present at 100%, whereas 7961C is present only at 20%, defining the minor haplogroup H1a1b, whereas 4639C and 10993A are present at 80%, defining the major haplogroup H1a1a1. (B) A mixture of two haplotypes within a single lineage but of different lineage depths (minor haplotype H1a1 and major haplotype H1a1a1) is observed if no minor haplotype can be found. (C) A mixture of two haplotypes within a single lineage but of different lineage depths (minor H1a1a1 and major H1a1) is detected if the minor haplotype results in a stable haplogroup. Shared homoplasmic sites facilitate the identification of the branching pattern in all three scenarios and improve the overall haplogroup quality. The used notation for variants (e.g., 1438G) includes the mtDNA position (1438) followed by the actual base change (G).











