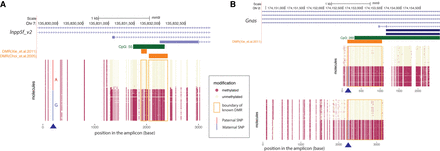
Phasing of 5mC with heterozygous variants using LR-EM-seq. (A) Phasing of 5mC with SNP of a 3.1-kb region in the imprinted Inpp5f_v2 gene promoter of the mouse cortex brain from a F1 offspring of a cross between two inbred mouse strains (129X1/SvJ male and Cast/EiJ female). Methylation state of individual CpG sites at the single-molecule level is denoted by either a beige dot (unmodified) or a red dot (methylated). The heterozygous SNP near the 5′ end of the region was either highlighted in red for paternal allele (A) or blue for maternal allele (G). The orange boxes denote previously identified DMRs. Our result not only confirmed the existence of the imprinted DMR but also revealed much extended boundaries of the imprinted DMR. (B) Phasing of 5mC and SNP in the imprinted promoter of the Gnas gene in the mouse cortex from a cross between the inbred mouse strains 129X1/SvJ (male) and Cast/EiJ (female). Methylation state of individual CpG sites at single-molecule level is denoted by either a beige dot (unmodified) or a red dot (methylated). The heterozygous SNP was highlighted in red for paternal allele (A) and blue for maternal allele (G). The orange box denotes a previously identified DMR. Our result confirmed the existence of the imprinted DMR and further extended this DMR in both directions particularly into the CpG island.











