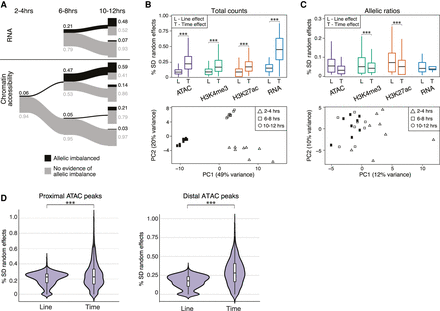
AI is generally not predictive of developmental time. (A) The relationship of AI across time for RNA (top) and chromatin accessibility (bottom). Proportions of AI and non-AI features shown in black and gray, respectively, and represented by the thickness of line (exact numbers indicated). Data for RNA at 2–4 h are not included owing to presence of maternal transcripts. (B, top) Box plots show distribution of time (T) and line (L) effect sizes obtained from mixed linear models for total counts. (Bottom) Principal component analysis (PCA) of gene expression for total counts. (C, top) Box plots showing the distribution of time (T) and line (L) effect sizes obtained from mixed linear models for allelic ratios. (Bottom) PCA of gene expression for allelic ratios. (D) Results from mixed linear models examining the effect of developmental time versus line (genotype) between proximal and distal ATAC-seq peaks for total count data. Distal peaks show a larger time effect compared with genotype effect (Mann–Whitney U test, P < 2.2 × 10−61). This is only slightly evident for promoter-proximal peaks (Mann–Whitney U test, P < 8.5 × 10−5).











