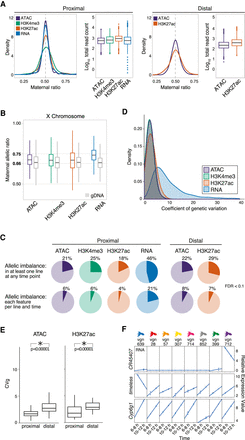
Allelic imbalance (AI) is common across regulatory data types. (A) Density plot of allelic count distribution for representative cross (vgn × DGRP-028) at 10–12 h and matching box plots showing total read count abundance (log10) in autosomes at TSS proximal (left) and distal (right) regions. Density plots for all crosses shown in Supplemental Figure S2B. (B) Box plot shows the distribution of maternal allelic ratios on the X Chromosome compared with genomic DNA (gray). (C) Pie charts show AI genes/features at promoter-proximal (left; TSS ±500 bp) and promoter-distal (right; 500–1500 bp ± from TSS) regions for all data types (FDR < 0.1). (Top) AI events in at least one F1 line at any time point. (Bottom) AI events detected in all eight F1 lines in all time points, on a per line and time basis. (D) Smoothed histograms showing distribution of coefficients of genetic variation for all features with significant between-line variances within each regulatory layer. (E) Box plots show the distribution of coefficient of genetic variation (CVg; y-axis) for chromatin accessibility (left) and H3K27ac signal (right), for promoter-proximal and promoter-distal sites. Genetic influences are more pronounced at distal elements in ATAC and H3K27ac. (F) Three examples of individual lines having distinct expression variation on specific genes. Relative expression values are typically larger for RNA than noncoding features, an effect that often results from one or two lines as shown here.











