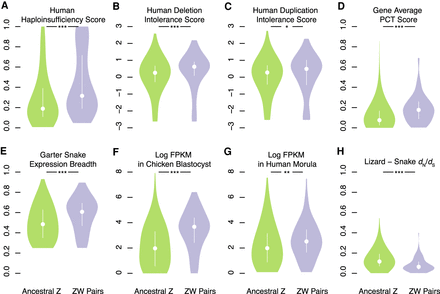
Factors in the survival of caenophidian Z–W gene pairs. Violin plots, with the median (white circle) and interquartile range (white bar) indicated, compare annotations of ancestral Z–W gene pairs identified in three species (purple) to annotations for the remainder of ancestral Z genes (green). (*) P < 0.05; (**) P < 0.01; (***) P < 0.001. P-values were obtained using one-tailed Wilcoxon rank-sum tests (Methods; Supplemental Table S24). Human orthologs of ancestral caenophidian Z–W gene pairs have greater probability of haploinsufficiency (A), deletion intolerance scores (B), duplication intolerance scores (C), and mean probabilities of conserved targeting (PCT) (D) than other ancestral Z genes. Orthologs of ancestral Z–W gene pairs are more broadly expressed than orthologs of other ancestral Z genes in a panel of seven adult eastern garter snake tissues (E). Orthologs of ancestral Z–W gene pairs are more highly expressed than orthologs of other ancestral Z genes in chicken blastocysts (F) and in human preimplantaion embryos (G). Orthologs of ancestral Z–W gene pairs have reduced dN/dS ratios compared to orthologs of other ancestral Z genes in alignments between tiger snake and green anole lizard orthologs (H).











