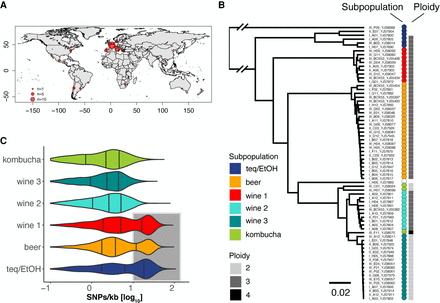
Ploidy and intra-genomic variation. (A) Strain collection. The 71 sequenced strains come from the collection of 1500 isolates (Avramova et al. 2018) and were isolated in different regions worldwide, where they are associated with anthropized environments such as tequila/bioethanol, beer, wine, and kombucha production. (B) Genetic relationship and ploidy level. The sequenced strains, here clustered based on Illumina short-read sequencing data (75PE), separate into six genetically distinct subpopulations—namely, tequila/bioethanol (teq/EtOH), beer, wine (1–3), and kombucha (based on 24,313 genome-wide distributed variants). Forty-eight strains were detected as triploids (69%) coming from five of the six subpopulations: teq/EtOH, beer, wine (1,2), and kombucha (inferred from genome-wide allele frequencies). (C) Genetic diversity within clades inferred from long-read sequencing data. The three subpopulations teq/EtOH (n = 5), beer (n = 22), and wine 1 (n = 7) harbor strains with two clusters of reads bearing low and high genetic variation (underlaid in gray) compared to the reference genome Brettanomyces bruxellensis (Fournier et al. 2017). The subpopulation wine 2 (n = 9), although being polyploid (B), lacks genomic regions with high genetic variation to the reference genome. The three lines within each distribution show the 25%, 50%, and 75% quartiles.











