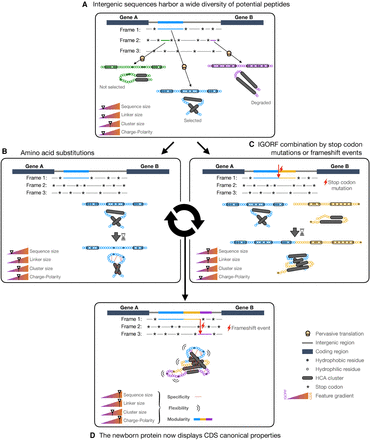
Model of de novo gene emergence and protein evolution with IGORFs as elementary structural modules. (A) IGORFs encode a wide diversity of peptides from disorder-prone to aggregation-prone ones, among which, a vast amount is expected to be able to fold in solution. Upon pervasive translation, some peptides that can be deleterious or not will be degraded right away. Among the others, the blue one will confer an advantage to the organism and will be further selected, thus providing a starting point for de novo gene birth. (B) The starting point IGORF, once selected, is subjected to amino acid substitutions, thereby increasing the overall proportion of hydrophilic residues of the encoded peptide. In the present case, this induces (1) the disruption of the second cluster, resulting in the increase of the size of the central linker, and (2) the establishment of specific interactions between hydrophilic residue (red dots), which increase the specificity of the folding process and the resulting fold. (C) The STOP codon of the starting point IGORF can be mutated into an amino acid, thereby adding the yellow IGORF to the pre-existing selected IGORF and elongating its size. (D) After multiple events of amino acid substitutions and IGORF combinations through STOP codon mutations or indels, we obtain a protein that displays the canonical features of CDSs (i.e., long sequences, long linkers, enrichment in polar and charged residues), which enable the optimization of its flexibility and the increase in specificity of its folding process, 3D fold, and interactions and finally participate along with domain shuffling or duplication events in the modular architecture of genuine proteins. We note that although the figure focuses on de novo gene emergence, this model can also apply to already existing proteins.











