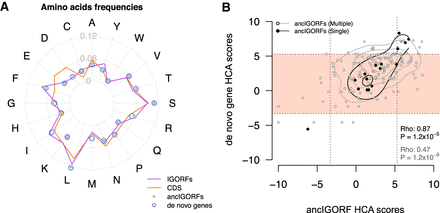
Impact of amino acid substitutions and IGORF fusion on the foldability of de novo genes. (A) Radar plot reflecting the 20 amino acid frequencies of IGORFs, ancIGORFs, de novo genes, and CDSs. (B) Plot of the HCA score of each de novo gene with those of its parent ancIGORF(s). The fold potential of a single-ancIGORF de novo gene is mostly determined by the one of its parent ancIGORFs, whereas the combination of several ancIGORFs through indels and STOP codon mutations leads most of the time to a foldable product. Single- and multiple-ancIGORF de novo genes are represented by black and white points, respectively. Spearman's correlation coefficients of the relationships between single- and multiple-ancIGORF de novo genes’ HCA scores versus the score of their parent ancIGORF(s), as well as the corresponding P-values, are indicated on the plot. The contour lines mark the percentiles of the density function range in black and gray for single- and multiple-ancIGORF de novo genes, respectively. The light pink region indicates de novo genes encoding proteins predicted as foldable.











