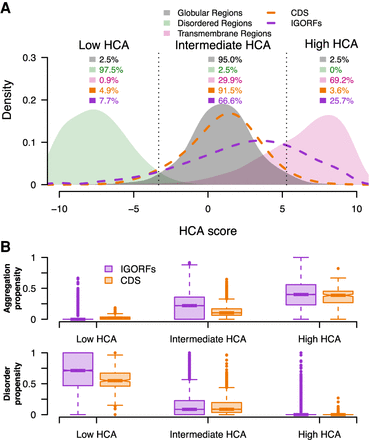
IGORFs encompass the large spectrum of fold potential of canonical proteins. (A) Distribution of the HCA scores for the three reference data sets (i.e., disordered regions, globular domains, and transmembrane regions; green, black, and pink curves, respectively) along with those for the CDSs (orange curve) and IGORFs (purple curve). There is a clear distinction between the distributions of HCA scores calculated for the three reference data sets (two-sided Kolmogorov–Smirnov test, P < 2 × 10−16 for all comparisons). Dotted black lines delineate the boundaries of the low, intermediate, and high HCA score categories, reflecting the three categories of fold potential (i.e., disorder prone, foldable, or aggregation-prone in solution). The boundaries are defined so that 95% of globular domains fall into the intermediate HCA score category, whereas the low and high HCA score categories include all sequences with HCA values that are lower or higher than those of 97.5% of globular domains, respectively. High HCA scores reflect sequences with high densities in HCA clusters that are likely to form aggregates in solution. Low HCA scores indicate sequences with high propensities for disorder, whereas intermediate scores correspond to globular proteins characterized by an equilibrium of hydrophobic and hydrophilic residues (Methods). The percentages of sequences in each category are given for all data sets. Raw data distributions are presented in Supplemental Figure S6. (B) Aggregation and disorder propensities calculated with TANGO and IUPred, respectively, are given for CDSs and IGORFs of each foldability HCA score category.











