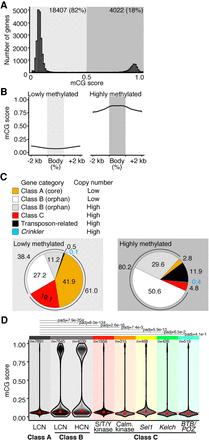
Methylation state of genes. (A) Distribution of methylation levels of R. irregularis genes. (B) Metagene plots of mCG methylation across genes with low (left) or high (right) mCG scores and their 2-kb upstream and downstream sequences. (C) Protein domain predictions of lowly and highly methylated genes. Pie charts show percentage of genes of each class, as categorized by the identity of their protein domains. Core (Class A) are nonrepeated genes with an identifiable protein domain. Class B genes contain no known protein domain. The class C gene category includes serine/threonine/tyrosine (S/T/Y) kinase, calmodulin-dependent kinase, Sel1-like, Kelch-like, and BTB/POZ. Transposon-related genes contain domains such as reverse transcriptase. Crinkler-type genes have a crinkler domain. (D) mCG score distribution of Class A (low copy number; LCN), Class B genes (high/low copy number; HCN/LCN), and high copy number Class C genes (rainbow-colored). A Kruskal–Wallis Dunn's multiple comparisons test (Benjamini–Hochberg correction) comparing mCG score distributions of gene groups was used to assess significance. Only adjusted P-values for comparisons to Class A are shown. The dotted red line highlights Class A median mCG levels.











