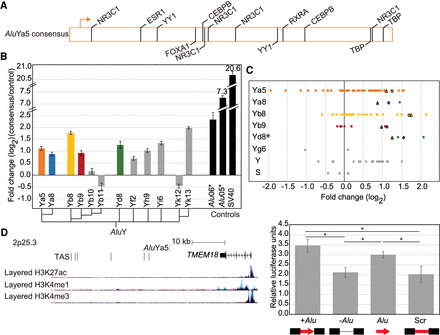
Alu elements have intrinsic regulatory potential. (A) AluYa5 has 14 high confidence putative transcription factor binding sites. (B) Ectopic luciferase assay results with isolated Alu consensus sequences. Cladogram shows the approximate evolutionary relationship between the 12 commonly polymorphic AluY subfamily consensus sequences, including five (colored) represented in our evaluated polymorphic Alu loci. Ectopic assay results for non-polymorphic, evolutionarily older Alu elements (*) previously evaluated in Su et al. (2014), and the strong SV40 enhancer are also shown (black). (C) Results from Figure 1B separated based on the Alu subfamily present at each locus. In cases where the consensus sequence was evaluated in B (matching colors), results for the consensus are shown as triangles (Alu tested sense with respect to luciferase) and diamonds (Alu consensus tested antisense with respect to luciferase). Alu subfamily does not drive the locus-specific results, as only the AluYd8 subfamily gave consistently distinct results (*) P = 0.02, ANOVA. (D, left) Genomic locus for Alu-609 drawn to scale with annotated epigenetic marks as in Figure 3. The AluYa5 (red) does not map to any notable epigenetic marks. This region was identified in GWAS as associated with obesity and body mass index (GWAS trait-associated SNPs [TAS]). Right, luciferase assay results for the locus with (+) and without (−) the Alu present compared to the isolated Alu-609 sequence (Alu). (*) Adjusted P < 0.05, t-test. Error bars are the standard deviation of two clones tested in triplicate in two experiments (n = 12).











