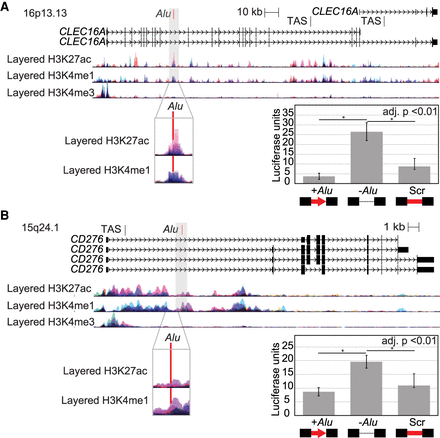
Polymorphic Alu elements mapping to epigenetic marks disrupt other genomic regulators. Genomic loci drawn to scale with layered chromatin marks from ENCODE tracks on UCSC Genome Browser and trait-associated SNPs (TAS) annotated. A magnified view (gray box) highlights the location of the polymorphic Alu (red) relative to chromatin marks. Ectopic luciferase reporter assay results are shown as relative luciferase units for each construct relative to a control vector. Comparisons were made between the locus with the Alu, without any insert, or with a scrambled Alu sequence (Scr) (t-test, adjusted for three comparisons). Error bars are the standard deviation of two clones tested in triplicate in two experiments (n = 12). (A) Polymorphic Alu element mapping to intron of CLEC16A, a region associated with type 1 diabetes. (B) Polymorphic Alu element mapping to an intron of CD276, a region linked to liver enzyme levels.











