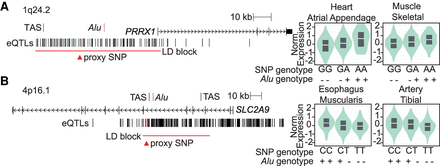
Polymorphic Alu outliers in the enhancer assay are associated with known eQTLs. Two example loci are shown, drawn to scale, with GWAS trait-associated SNPs (TAS) and Alu element (red) location marked. GTEx identified eQTLs, including the SNP used as a proxy for the polymorphic Alu element (red triangle), are annotated as well as the extent of the linkage disequilibrium (LD) surrounding the proxy SNP (red bar). The presence (+) or absence (−) of the Alu was phased with the proxy SNP genotype, and GTEx genotype-dependent expression is shown for two example tissues. (A) Polymorphic Alu at PRRX1 is candidate causative variant in atrial fibrillation risk GWAS (TAS = rs39033239, r2 = 0.4, D′ = 0.93). A GTEx eQTL, rs10489231, is a perfect proxy for the Alu (r2 = 1). (B) Polymorphic Alu at SLC2A9 maps to uric acid and gout GWAS signals (TAS = rs3775948 and rs4475146). GTEx eQTL rs4235346 is a perfect proxy for the Alu (r2 = 1).











