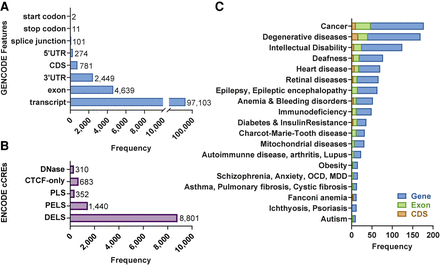
MEI mutagenesis patterns in GENCODE, ENCODE, and various databases. (A) Comparisons of the combined collection of 158,783 MEIs depicted in Figure 1B revealed intersections with GENCODE v35 gene annotations. The total number of GENCODE transcripts intersected by MEIs is 97,103. All features impacted by MEIs (exons, etc.) are found within these transcripts. The UTRs and CDSs also are included in the exon group, because they also are exons. In cases in which multiple transcript models are intersected by MEIs, all transcripts and features are listed in Supplemental Table S6A and are delimited by commas in the same order for each column. We identified insertions that disrupt various subregions of genes including 781 MEIs that disrupt CDS exon sequences. (B) MEIs disrupt ENCODE-annotated cis-candidate regulatory elements (cCREs). (C) MEIs disrupt genes that have been linked to various diseases in the Online Mendelian in Man (OMIM) database. Although all these MEIs disrupt genes and their annotated features, these insertions may or may not have functional consequences. MEIs that disrupt coding exons or ENCODE transcriptional regulators likely produce functional consequences. MEIs that occur within introns, UTRs, and other functionally important sites also can impact gene function (e.g., Watanabe et al. 2005; Lanikova et al. 2013). However, the precise functional consequences of gene-disrupting MEI insertions can be difficult to predict and must be validated experimentally.











