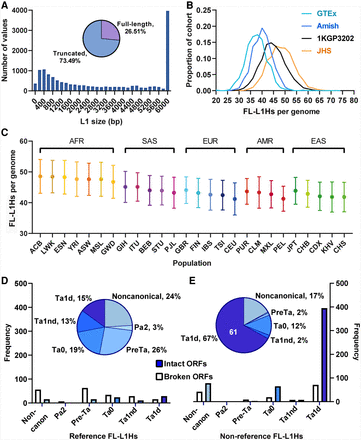
FL-L1Hs elements discovered in this study. (A) We identified 3728 FL-L1Hs elements that are 6 kb or longer within the collection of 16,525 L1 MEIs that were discovered in this study. Because MELT could estimate the lengths of 14,066/16,525 (85.1%) L1s, we calculated the percentages in A using the denominator of 14,066. (B) Comparisons of FL-L1Hs elements per genome in the four WGS studies of our study. The GTEx and Amish populations have fewer FL-L1Hs copy numbers compared to the 1KG population and the Jackson Heart Study. (C) Population distribution of FL-L1Hs elements arranged by superpopulation of the 1KGP 2504 high-coverage genomes. (D,E) REF and non-REF FL-L1Hs elements with their subfamily distributions. Corresponding bar plots indicate the number of FL-L1Hs elements with two intact ORFs (solid bars). (E) The majority of FL-L1Hs elements in the non-REF group belong to the Ta1d subfamily and have two intact ORFs (bottom right). Note also the expansion of Ta1d elements in non-REF populations compared to REF (from 15% to 67%; compare purple sections in D and E). The white 61 numeral in E indicates the number of elements in this group with documented activity in the literature (Supplemental Table S3A).











