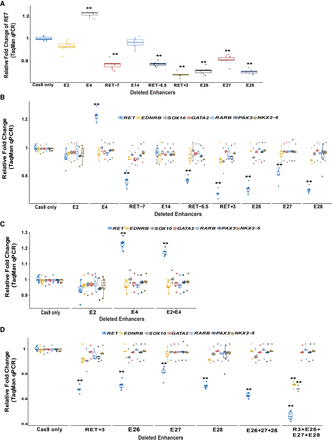
CRISPR-Cas9-induced deletions of RET enhancers with HSCR-associated variants reduce RET gene expression in vitro. (A) There is significant loss of RET gene expression from seven of nine CREs with small (≤10-bp) deletions centered on the variant site; only the E4 enhancer shows increased gene expression. (B) The expression of other RET GRN genes is unaffected by these CRE deletions likely owing to RET gene expression not decreasing below 50%. (C) Simultaneous deletion of enhancers E2 and E4 does not lead to any additional effect on RET gene expression compared with individual deletions. (D) Simultaneous deletions of E26/E27/E28 with and without deletion of RET+3 lead to a significantly greater decrease in RET gene expression compared with individual deletions. The deletion of all four enhancers also leads to decreased expression of EDNRB and SOX10. (**) P < 0.001 in two technical replicates each for three independent biological replicates in all experiments.











