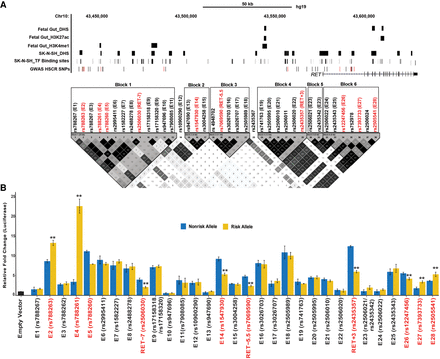
The RET regulatory landscape in the enteric nervous system. (A) The 155-kb RET locus (Chr 10: 43,434,933–43,590,368; hg19) contains 38 HSCR-associated polymorphisms in six linkage disequilibrium (LD) blocks. LD between all 38 SNPs was estimated as by Gabriel et al. (2002). Multiple enhancer-associated epigenetic marks (DNase I hypersensitivity [DHS], H3K27ac, H3K4me1) in 108-day human fetal large intestine and the SK-N-SH neuroblastoma cell line and transcription factor (TF) binding sites (TFBSs) from public sources are noted. All common (minor allele frequency ≥10%) variants associated with HSCR are shown, with those showing allelic difference in in vitro transcription assays marked in red. (B) Allele-specific in vitro luciferase assays of 28 CREs containing 30 polymorphisms plus three previously tested controls in SK-N-SH cells are shown: 22 CREs act as enhancers, compared with a promoter-only control, of which seven also show allelic difference in luciferase activity (boxed) for its cognate HSCR-associated polymorphism in addition to RET-7, RET-5.5, and RET+3 positive controls. Error bars are SEM of three independent biological replicates: (**) P < 0.001.











