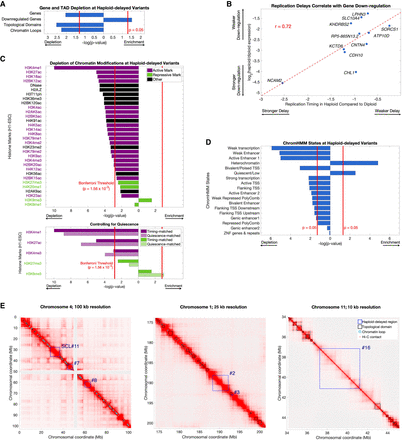
Haploid-delayed variants are located in regions of unorganized, quiescent heterochromatin depleted of genes, histone marks, and 3D contacts. (A) Haploid-delayed variants show a significant depletion of genes, topologically-associated domains (TADs), and chromatin loops, and an enrichment of genes down-regulated in haploid ESCs. Bars indicate enrichment/depletion two-sided t-test P-value of each tested feature in replication timing variants compared to 1000 matched permutations. (B) The extent of haploid replication delay correlates with the magnitude of gene expression differences between haploid and diploid cells. Spearman's rank correlation was similar (rho = 0.65); thus, this correlation is not driven by NCAM2. (C) Haploid-delayed variants are depleted of chromatin marks. Bars shown as in A. Each chromatin mark was considered to be independent, and the Bonferroni-corrected P-value 0.0016 (0.05/32) is shown as a solid red line. Histone mark activation/repression information was obtained from Schiltz et al. (1999) and Zhao and Garcia (2015). Bottom: Three of five tested histone marks (chosen due to their similarity with UR regions) remain depleted when controlling for quiescence, whereas H3K27me3 remained nominally depleted, and H3K9me3 became significantly enriched. P-values for matched permutations (dark bars) are identical to the upper panel, with P-values for corresponding quiescence-matched permutations (light bars) shown below them. (D) As in A and C, for ChromHMM states. Overlap was calculated by the number of base pairs of each state at variant regions. ChromHMM states are mutually exclusive (i.e., not independent); thus, a P-value cutoff of 0.05 was used for significance. (E) Hi-C chromosome interaction maps in H1-ESC (Dekker et al. 2017) at three different genomic locations and resolutions. Haploid-delayed variants (dark blue) have little overlap with chromosome domains (black) or loops (light blue). This trend is observed across all haploid-delayed variants (panel A).











